An Okazaki Fragment Has Which of the Following Components
Figure 1421 A fr am eshift mutation that results in the insertion of three nucleotides is often. This requirement has two fundamental consequences.
Biomolecules Free Full Text Primpol A Breakthrough Among Dna Replication Enzymes And A Potential New Target For Cancer Therapy Html
All 5 and 3 ends b helicase f DNA polymerase III Leading strand c DNA ligase g primase Lagging strand d RNA primer Okazaki fragment e single-strand binding proteins a DNA polymerasel மடடபப Below is a picture of a single chromosome that was duplicated.

. Thus each primer originates at or near the replication fork and is extended in the opposite direction. An Okazaki fragment has which of the following arrangements. This is a Most important question of gk exam.
The large fragment known as the Klenow fragment is widely used for DNA sequencing and many other techniques that require DNA synthesis without 5 3 degradation. 1 The lagging strand must have evolved priming and fragment joining mechanisms involving many additional steps and. What are the okazaki fragments.
5 DNA to 3. Solid boxes represent the primers and dotted lines represent the newly synthesized DNA strands. The reaction buffer was 10 mM magnesium acetate 70 mM KCl 50 mM Hepes pH 75 100 mM potassium glutamate 20 glycerol 200 µgml bovine serum albumin 002 Nonidet P-40 and 10 mM dithiothreitol.
A RNA nucleotides DNA nucleotides b DNA polymerase I DNA polymerase III c RNA primer DNA fragment d Primase polymerase ligase. The strand is synthesized in short segments named Okazaki fragments after their discoverer Sakabe and Okazaki 1966. Do you prefer DNA polymerase 1 or 3.
An Okazaki fragment has which of the following arrangements. Primase polymerase ligase O d. Then DNA polymerase III binds to the RNA primer and adds deoxyribonucleotides as short segments Okazaki fragments.
What binds Okazaki fragments together. DNA synthesis is continuous on the leading strand. Options is.
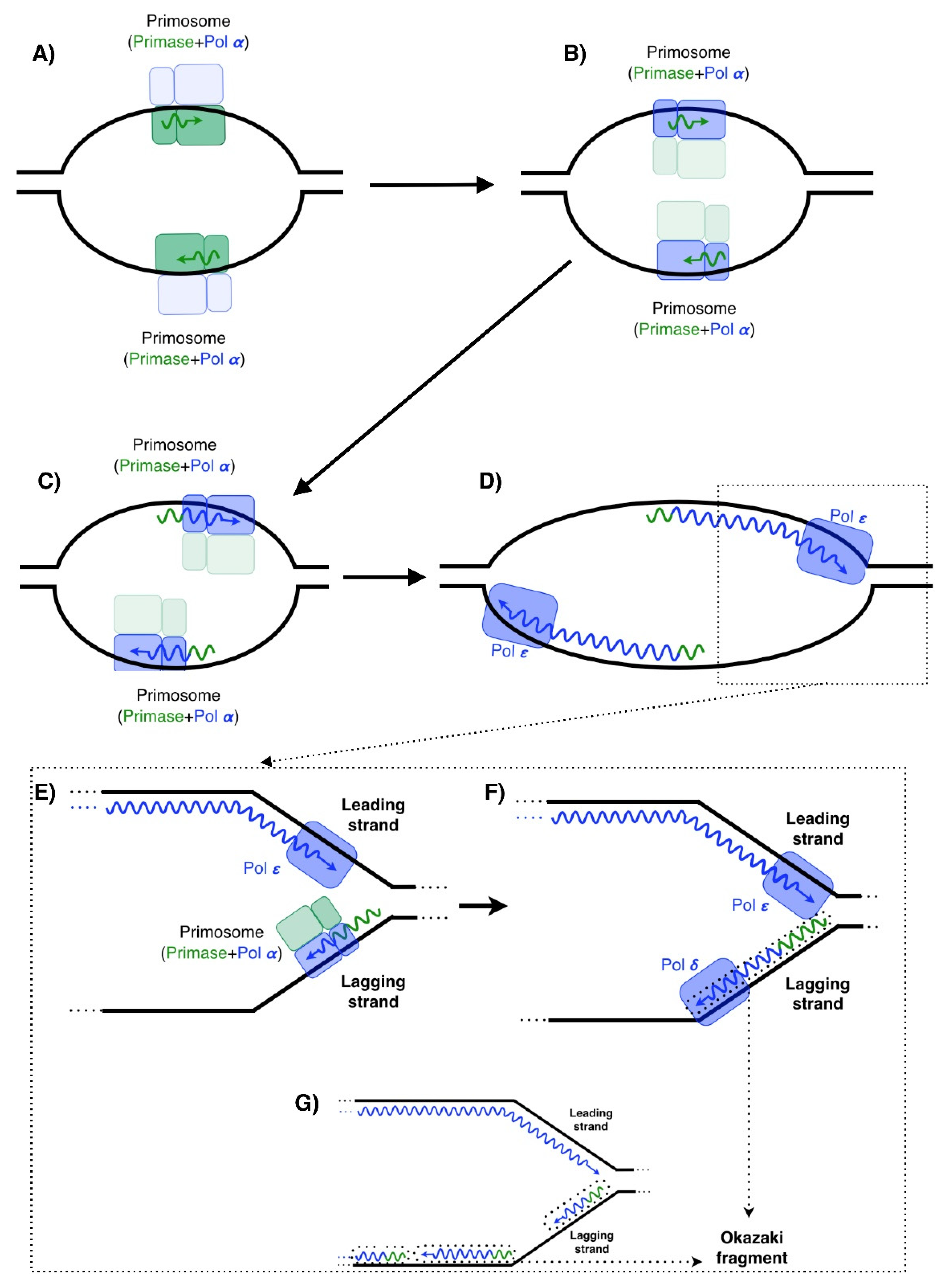
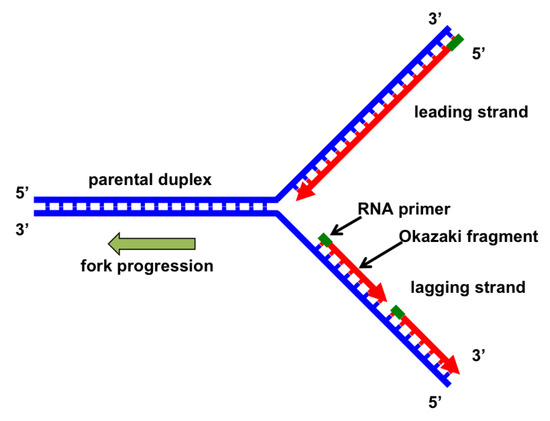
For the synthesis of the lagging strand first an RNA primer is created. During replication of DNAwhich one of the following enzymes polymerizes the Okazaki fragments. The primer for each new Okazaki fragment is synthesized in the 5 to 3 direction by primase a DNA-dependent RNA polymerase which is also component of the primosome along with helicase and other DNA binding proteins Fig.
PubMed Google Scholar Bocquier AA. All were diluted in KCl-EDB running buffer. Figure 1414 You isolate a cell strain in which the joining of Okazaki fragments is impaired and.
If DNA of a particular species was analyzed and it was found that it contains 27 percent A what. It was used to adjust the length of Okazaki fragments to the physiological range of 12 kb. DNA Polymerase I 3RNA Polymerase I 4.
The isolation of nascent Okazaki fragments during DNA replication led to the surprising. DNA polymerase I DNA polymerase II E. An okazaki fragment has which of the following components.
1968 and the segments are then joined. An Okazaki fragment has which of the following arrangements. Primase polymerase ligase B.
He hypothesized that in this situation DNA polymerase would quit its job once it ran out of space and then swing back to. They were named after Reiji Okazaki who using radioactive labels in bacterial DNA observed an increase of short oligonucleotides directly after the replication process had begun but a. A primase polymerase ligase B 3 RNA nucleotides DNA nucleotides 5 C 5 RNA nucleotides DNA nucleotides 3 D DNA polymerase I DNA polymerase III E 5 DNA to 3 Start studying Bio Ch 16.
A B Template DNA a. This is where Reiji Okazaki comes into the picture. DNA contains which one of the following components.
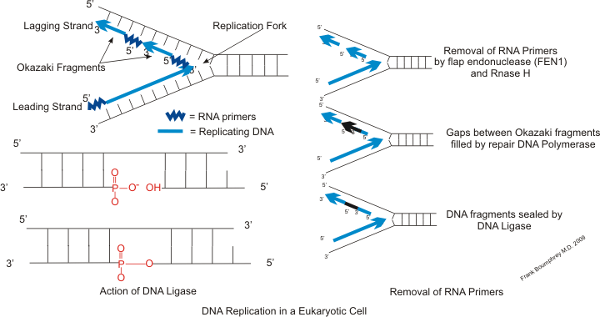
The term Okazaki fragment relates to short oligonucleotide sequences that are synthesised during DNA replication as part of the lagging strand of DNA running in the 5 3 direction. DNA polymerase I DNA polymerase III O e. After the removal of the RNA primers and their replacement with DNA the Okazaki fragments are joined together to form an unbroken strand of DNA that carries.
Whilst this canonical pathway for the maturation of Okazaki fragments is very efficient it has been estimated from biochemical experiments that 15-30 of human POLδ molecules disengage before. 5RNA nucleotides DNA nucleotides 3 b. An Okazaki fragment has which of the following components.
If a 1000-kilobase fragment of DNA has 10 evenly spaced and symmetric replication origins and DNA polymerase moves at 1 kilobase per second how many seconds will it take to produce two daughter. To which direction towards the left or. The following components were used either individually or in combination to determine the requirements for full recycling.
DNA Polymerase II 2. Label the following components of DNA replication in the diagram below. 3 RNA nucleotides DNA nucleotides 5 o c.
3RNA nucleotides DNA nucleotides 5 C. 13 An Okazaki fragment has which of the following arrangements. What is the purpose of okazaki fragments.
DNA Polymerase III 5. DNTPs 40 µM of each final ddNTPs 40 µM final ATP 1 mM final ATPγS 1 mM final β 2 05 µM final SSB 4 05 µM final 20-mer70-mer 05 µM final. On the lagging strand DNA synthesis restarts many times as the helix unwinds resulting in many short fragments called Okazaki fragments DNA ligase joins the Okazaki fragments together into a single DNA molecule.
5RNA nucleotides DNA nucleotides 3 D. In the following diagram A and B are two Okazaki fragments generated during DNA replication. 5 DNA to 3 5 RNA nucleotides DNA nucleotides 3 3 RNA nucleotides DNA nucleotides 5 DNA polymerase I DNA polymerase III primase polymerase ligase.
The larger fragment consists of the C-terminal 605 amino acid residues and the smaller fragment contains the remaining N-terminal 323 residues.

Dna Replication Leading Strand Vs Lagging Strand Okazaki Fragments Youtube

2 631 Dna Replication Stock Photos Pictures Royalty Free Images Istock

The Roles Of Family B And D Dna Polymerases In Thermococcus Species 9 N Okazaki Fragment Maturation Journal Of Biological Chemistry

Cdc9 Regulates The Fidelity Of Okazaki Fragment Maturation A Model Download Scientific Diagram

Caned Chromosome Chromosome En Boite Polymer Clay Necklace Clay Necklace Polymer Clay Beads

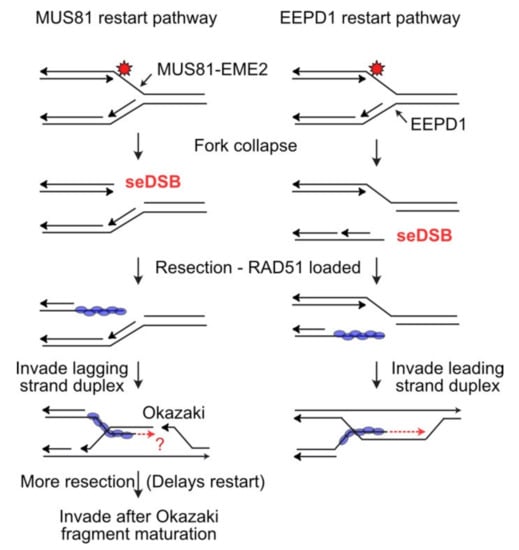
Eukaryotic Lagging Strand Dna Replication Employs A Multi Pathway Mechanism That Protects Genome Integrity Journal Of Biological Chemistry

Okazaki Fragments An Overview Sciencedirect Topics

Direct Visualization Of Rna Dna Primer Removal From Okazaki Fragments Provides Support For Flap Cleavage And Exonucleolytic Pathways In Eukaryotic Cells Journal Of Biological Chemistry

Okazaki Fragments An Overview Sciencedirect Topics

Dna Replication And Repair Google Slides And Distance Learning Ready Genetics Lesson Biology Lessons Science Lessons High School

Replication Fork Teaching Biology Study Biology Biology Classroom
True Regarding Okazaki Fragment Biochemistry Mcq

Southern Dna Probed By Antiparallel Dna Sequence Vs Northern Rna Probed By Complementary Rna V S Western Protein Dna Probe Biochemistry Molecular Biology

Dna Replication Catalyzed By Herpes Simplex Virus Type 1 Proteins Reveals Trombone Loops At The Fork Sciencedirect
Dna Free Full Text Nucleases And Co Factors In Dna Replication Stress Responses Html
Genes Free Full Text The Replication Fork Understanding The Eukaryotic Replication Machinery And The Challenges To Genome Duplication Html


Comments
Post a Comment